CUT&Tag
CUT&Tag (Cleavage Under Targets and Tagmentation) offers a novel approach to pursue epigenetics. The method is designed to map genome wide histone variants and post-translational modifications, transcription factor binding sites, and chromatin-associated complexes.
CUT&Tag Workflow
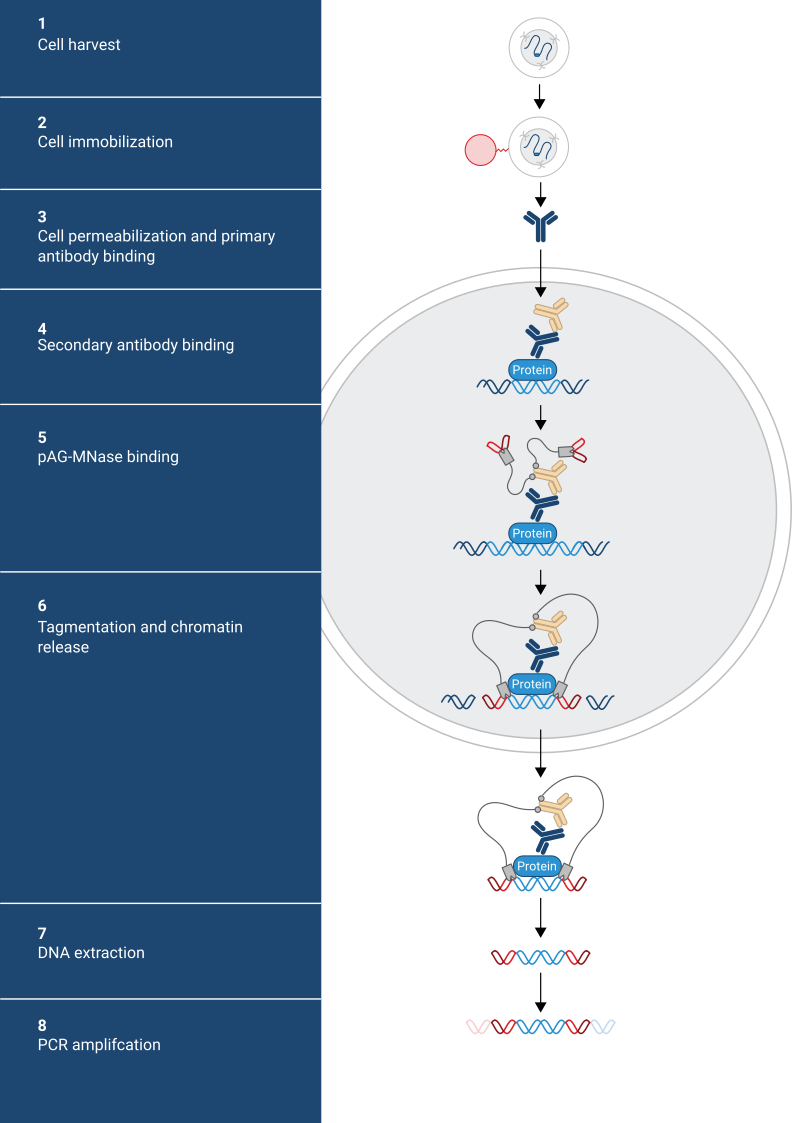
Steps of standard CUT&RUN protocol: 1. Cell harvest 2. Cell immobilization 3. Cell permeabilization and primary antibody binding. 4. Secondary antibody binding 5. pAG-MNase binding 6. Tagmentation and chromatin release 7. DNA Extraction 8. PCR amplification
Get our free CUT&RUN and CUT&Tag handbook now!
Show moreCUT&Tag is performed in situ on immobilized, intact cells without crosslinking. DNA fragmentation is achieved through tagmentation of genomic DNA by a hyperactive Tn5 transposase that is loaded with sequencing adapter duplexes and fused to Protein A and/or protein G (pA/G-Tn5). The fusion protein is tethered via its Protein A and/or Protein G moieties to an antibody against the desired target, thus leading to tagmentation in the vicinity of the targeted antigen. CUT&Tag products are suitable for library generation for next generation sequencing (NGS) without any further end polishing steps.
Advantages of CUT&Tag
- Performed In situ on non-fixed cells; no chromatin fragmentation necessary.
- Low background and high sensitivity require low sequencing depth.
- No end-polishing and sequencing adapter ligation steps necessary.
- Possible with low cell numbers down to 100 cells depending on the antigen.
- Simple, fast, amenable to automation.
- Accurate quantitation using carry-over E. coli DNA from the pAG-Tn5 purification.
Products for CUT&Tag
antibodies-online offers flexible solutions for your CUT&Tag experiments. Either pick yourself control antibodies, secondary antibodies or magnetic beads from our CUT&Tag product offer contact our team of biologist for a finely tuned set of reageants alongside with an optimizied protocol.
 豚鼠 anti-兔 IgG (Heavy & Light Chain) Antibody - Preadsorbed (ABIN101961)
豚鼠 anti-兔 IgG (Heavy & Light Chain) Antibody - Preadsorbed (ABIN101961)
- CUT&Tag secondary antibody
- Signal amplification in CUT&Tag
- Efficient pAG-Tn5 Fusion tethering
 Magnetic Concanavalin A Beads (Agarose) (ABIN6952467)
Magnetic Concanavalin A Beads (Agarose) (ABIN6952467)
- Suitable for CUT&Tag Assays
- Superior handling compared to silica ConA beads
- In stock, fast delivery
- Valdidated for CUT&RUN and CUT&tag
- Detects specifically mouse IgG (H&L)
- 3 PubMed Citations available
Epigenetic Antibodies for CUT&Tag
antibodies-online offers antibodies against abundant histone modifications and transcriptionsfactors. They are suitable as positive control and important to assess the success of the CUT&Tag method.
Frequently asked Questions
How do I choose between CUT&RUN and CUT&Tag?
In CUT&Tag, sequencing primers are being attached to the cleaved DNA and no additional annealing step is necessary. It works well for nucleosomal and tightly bound proteins. The method is also less likely to produce background signal due to DNA damage.
CUT&RUN on the other hand is preferable for transcription factors (complexes) or proteins less tightly bound to DNA. It also has a better resolution than CUT&Tag.
Is it possible to use CUT&Tag with plant tissue samples?
CUT&Tag on isolated plant cells has been described for profiling of H3K4me3 histone marks in different cotton tissue types using isolated nuclei14. Our positive control H3K27me3 H3K4me3 antibodies and the negative control guinea pig anti-rabbit IgG antibody as well as the ConA beads are suitable for use with plant samples.
Can I denature the proteins in the CUT&Tag product complex instead of a proteinase K treatment?
We recommend against this option: the DNA of interest is at his point present in a complex consisting of the DNA, the protein of interest, the corresponding antibody, and the pA/G-Tn5. Boiling this complex will likely precipitate the DNA together with denatured protein. This will also primarily affect the short CUT&Tag products and not larger DNA molecules, leading to a decreased signal to noise ratio in your library and potentially also reducing the library’s complexity. This effect is further exacerbated because of the lower melting temperature of these short molecules compared to the longer contaminating DNA molecules.
Which secondary antibodies can I use for CUT&Tag?
- (133)
- (3)
- (2)
Can I use AMPure CP beads to purify tagmentation products instead of phenol-chloroform extraction?
In the original publication describing the CUT&Tag method the authors mention the use of AMPure XP beads for the purification of the DNA subsequently to tagmentation and Proteinase K digest1. A potential issue is the carry-over of active Proteinase K, which can interfere with the downstream PCR amplification. Therefore, the authors recommend now the phenol-chloroform extraction to assure complete denaturation of Proteinase K15.
Why is the DNA yield so low?
CUT&Tag is performed using low cell numbers and the background signal is considerable lower than e.g. for ChIP. Due to these two factors the amount of recovered DNA is often times too low to be reliably measured based on a fluorometric assay or by capillary electrophoresis. PCR amplification of small CUT&RUN products, i.e. less than 50 bp, can be problematic and is therefore not an option.
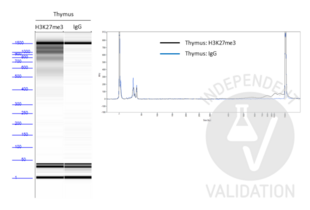
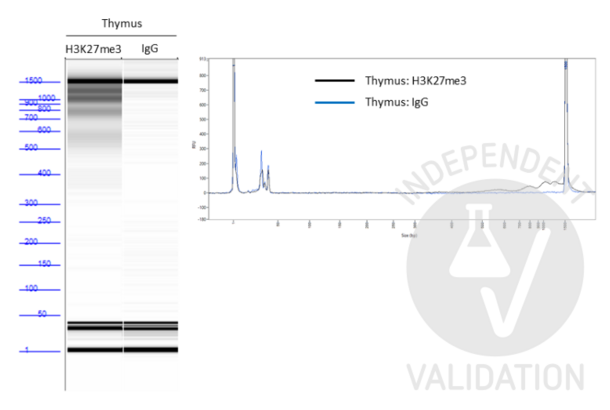
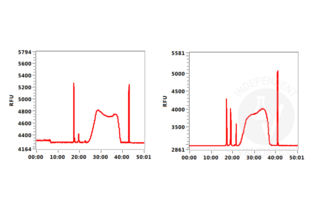
In order to assess the success of the CUT&Tag method include (a) positive control sample(s) using the Positive Control H3K4m3 Antibody ABIN3023254 and/or the Positive Control H3K27me3 Antibody ABIN6923144. DNA fragments prepared using these antibodies can be measured by sensitive electrophoresis on a Bioanalyzer or Tapestation or fluorometrically on a Qubit or Nanodrop fluorometer. When using the CUT&RUN Positive Control ABIN6923144 (red dot) (or any other antibody specific for nucleosomal markers) a ladder corresponding to multiples of the 147 bp long nucleosomes should be visible by capillary electrophoresis.
Related Information and Products
CUT&RUN - cleavage under targets and release using nuclease: Information and products
References
: "Efficient chromatin profiling of H3K4me3 modification in cotton using CUT&Tag." in: Plant methods, Vol. 16, pp. 120, (2020) (PubMed).: "Improved CUT&RUN chromatin profiling tools." in: eLife, Vol. 8, (2019) (PubMed).
: "The three-dimensional structure of a Tn5 transposase-related protein determined to 2.9-A resolution." in: The Journal of biological chemistry, Vol. 274, Issue 17, pp. 11904-13, (1999) (PubMed).
: "ChIC and ChEC; genomic mapping of chromatin proteins." in: Molecular cell, Vol. 16, Issue 1, pp. 147-57, (2004) (PubMed).
: "Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing." in: Science (New York, N.Y.), Vol. 348, Issue 6237, pp. 910-4, (2015) (PubMed).
: "An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites." in: eLife, Vol. 6, (2018) (PubMed).
: "CUT&Tag for efficient epigenomic profiling of small samples and single cells." in: Nature communications, Vol. 10, Issue 1, pp. 1930, (2019) (PubMed).
Henikoff, S. & Henikoff, J. G. Profiling the epigenome at home. bioRxiv 1–18 (2020). doi:10.1101/2020.04.15.043083
Bartosovic, M., Kabbe, M. & Castelo-Branco, G. Single-cell profiling of histone modifications in the mouse brain. bioRxiv 2020.09.02.279703 (2020). doi:10.1101/2020.09.02.279703
Wu, S. J. et al. Single-cell analysis of chromatin silencing programs in developmental and tumor progression. bioRxiv 2020.09.04.282418 (2020). doi:10.1101/2020.09.04.282418
Kaya-Okur, H., Henikoff, S. & Henikoff, S. Bench top CUT&Tag. protocols.io 1–19 (2019). Available at: https://www.protocols.io/view/bench-top-cut-amp-tag-z6hf9b6.

Creative mind of antibodies-online with a keen eye for details. Proficient in the field of life-science with a passion for plant biotechnology and clinical study design. Responsible for illustrated and written content at antibodies-online as well as supervision of the antibodies-online scholarship program.
前往作者页面



