COVID-19
Since the initial identification of the novel coronavirus SARS-CoV-2 as causative agent of COVID-19 almost 55.000.000 infections (as of Nov 2020) have been registered worldwide. Thanks to global research efforts, a picture of the virus biology and its effect in the human host is emerging. We offer a wide variety of high quality products for COVID-19 research.
Antibodies, Kits, Proteins by Disease Phase:
COVID-19 Products by Product Type:
SARS-CoV-2 Infection
SARS-CoV-2 primarily infection is initiated when the host cell angiotensin-converting enzyme 2 (ACE2) surface receptor is bound by the virus’ spike (S) protein through its receptor binding domain (RBD). ACE2 is encoded on the X chromosome, which might explain the higher COVID-19 fatality rate in men. Possibly, having two different ACE2 alleles confers some degree of resistance.
Binding of SARS-CoV-2 to ACE2 triggers priming of the trimeric S protein at the polybasic S1/S2 cleavage site by the cell surface-associated transmembrane protease serine 2 (TMPRSS2) and to a lesser degree cathepsin B and L. The S1 ectodomain containing the RBD determines cellular tropism and attachment of the virus to its target cell. The S2 endodomain harbors a transmembrane domain and is involved in virus entry through endocytosis. It also contains a second protease site, the furin-like S2’ cleavage site. Precleavage at this furin-like cleavage site might explain the higher infectivity of SARS-CoV-2 compared to SARS-CoV (which lacks this site). Cleavage of the S protein does also generate a C-end rule (CendR) motif at the C-terminus of the S1 ectodomain. This motif binds directly to the tyrosine kinase receptor Neuropilin-1 (NRP1), thus facilitating SARS-CoV-2 entry inot the cell.
Featured COVID-19 related Products:
- (8)
- (15)
- (1)
- (4)
- (5)
- (9)
- (8)
- (5)
- (3)
- (4)
- (3)
- (1)
Viral Replication
Upon entry, the viral positive-sense ssRNA(+) genome is released. Two large polycistronic open reading frames ORF1a and ORF1b at the 5’ end of the genome encode 16 non-structural proteins (NSPs) forming two replicase polyproteins pp1a and pp1b. Nsp3 contains a papain-like protease (PLpro) domain and processes Nsp1-4 of pp1a. The 3C-like main chymotrypsin-like protease (Mpro, 3CLpro, Nsp5) of SARS-CoV-2 digests the remaining proteolytic cleavage sites.
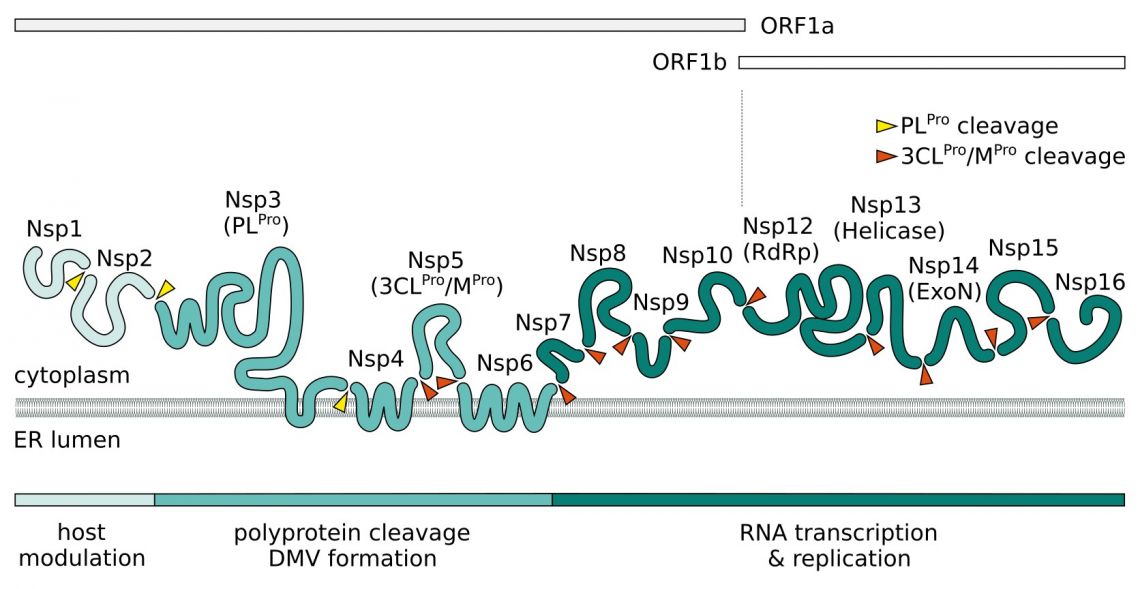
Nsp1 and Nsp2 are thought to play a role in host modulation and interefere with the antiviral response. SARS-CoV-2 Nsp1 suppresses the interferon pathway by blocking the host cell's ribosome mRNA entry channel. The result is an imbalanced immune response which propagates cytokine release and inflammation. A complex consisting of transmembrane proteins Nsp3, Nsp4, and Nsp6 induces formation of double-membrane vesicles (DMV) improving viral replication through membrane associated replication and affecting autophagy. In addition to its NSPs SARS-CoV-2 recruits host proteins to form a replication and transcription complexes (RTC). Core component for the replication of the ssRNA(+) is the RNA-dependent RNA polymerase (RdRp). It forms the replication complex together with Nsp7 and Nsp8. These serve as primase and generate short RNA primers for the primer-dependent RdRp and increase its processivity. Nsp9 has a preference for ssRNA and is believed to interact with Nsp8 in the replication complex. In SARS-CoV-2, both protein bind to the signal recognition particle that helps transport newly translated host proteins within the cell. Nsp13 and Nsp16/Nsp10 have helicase/triphosphatase and methyltransferase activity respectively and cap the nascent viral mRNA. Nsp16 also disrupts host mRNA splicing through binding to the mRNA recognition domains of U1/U2 snRNAs. The exonuclease Nsp14 (ExoN) endows the replication machinery with a proofreading function, thus increasing fidelity of SARS-CoV-2 RNA synthesis. The last protein of the replication complex is the uridine-specific endoribonuclease Nsp15 (EndoU).
Once the RTC is assembled, a dsRNA intermediate is synthesized from the genomic ssRNA(+). This intermediate serves as template for the production of subgenome-length RNAs (sgRNA) and new, full-length ssRNA(+) genome. The former are transcribed into the virus’ four structural proteins (N, E, M, and S) and nine accessory proteins encoded in the 3’ section of the genome. The latter is packaged into nucleocapsid phosphoproteins (N protein) and then enveloped by the envelope protein (E protein), the membrane glycoprotein (M protein), and spike protein (S protein) to form new virions.
Featured COVID-19 related Antibody Products:
- (8)
- (15)
- (1)
- (9)
- (8)
- (1)
- (2)
Featured COVID-19 related Protein Products:
- (1)
COVID-19
Disease severity in patients is due not only to the viral infection but also the host response. The host’s inflammatory response strongly influences the damage to the airways. In 70% of the fatal COVID-19 cases, the resulting acute respiratory distress syndromes (ARDS) leads directly to respiratory failure. The second-most common cause for fatalities in context with COVID-19 is an uncontrolled systemic inflammatory response driven by overproduction of inflammation markers. This so-called cytokine release syndrome or cytokine storm is an important contributor to ARDS and multiple organ dysfunction syndrome (MODS) causing damage in particular to heart, kidneys, and liver.
SARS-CoV-2 Infection and Inflammatory Response
SARS-CoV-2 primarily replicates in the lower respiratory tract where it causes pneumonia and ARDS. While the virus’ structural and non-structural proteins are mainly tasked with building up the virion and virus replication respectively, at least some of the members of the third group of SARS-CoV-2 proteins, the nine accessory factors (Orf3a-10), have been implicated in driving progression of COVID-19.
SARS-CoV-2 Orf3a induces apoptosis in cell line models and is thought to activate NF-kB and the NLRP3 inflammasome involved in pyroptosis, a highly inflammatory form of apoptosis. Orf8b was shown to induce ER stress and to also activate the NLRP3 inflammasome, as was the SARS-CoV-2 E protein. This suggests that viral infection of airway epithelial cells leads to pyroptosis. Pyroptosis is typically accompanied by the release of proinflammatory cytokines leading to the recruitment of additional immune cells and further amplification of the immune response. In addition, caspase-1 in the NLRP3 inflammasome processes the pro-forms of IL-1 beta and IL-18 into the active forms. Orf7a may also play a role in pathogenesis via its role in virus-induced apoptosis.
The extent of damage to tissue in the lower respiratory tract can be monitored in the early stages of COVID-19 using C-reactive protein (CRP) levels as an indicator for disease severity.
The destruction of the alveolar epithelial cells sets damage-associated molecular patterns (DAMPs) and pathogen‐associated molecular patterns (PAMPs) free, which are detected by pattern‐recognition receptors (PRRs) on alveolar epithelial cells and macrophages. The primary PRRs for viral RNAs are members of the RIG‐I‐like receptor (RLR) family. Upon binding to viral RNA, a conformational change of the RLRs triggers aggregation of MAVS and formation of the MAVS signalosome. The MAVS signalosome triggers IRF3/7 dimerization and activates NF‐κB pathway, leading to the production of type I IFNs, the most important antiviral cytokines, and pro‐inflammatory cytokines IL-6, IFN-gamma, CD46, and CXCL10. The coordinated secretion of pro-inflammatory cytokines and chemokines leads to recruitment of immune cells, in particular CD4+ T helper cells (TH1), CD8+ cytotoxic T cells and monocytes, to mount the defense against the viral infection.
As a type I IFN antagonist, SARS-Cov-2 Orf6 inhibits the IFN response. Orf9b targets the MAVS signalosome for degradation and therefore limits the host cell interferon responses. Orf9c interacts with the mitochondrial electron transport chain which is involved in TLR/IL-1 signaling and regulation of inflammation. Nsp1 suppresses IFN induction and increases CCL5 production, thus contributing to inflammatory processes.
SARS-CoV-2 Interferon Antagonism: Proteins & Antibodies
SARS-CoV-2 Cytokine Storm: Antibodies & ELISA Kits
Related Antibodies: CRP, NFKB1, NFKB2, NFkB cRel, p65, RELB
- (2)
- (7)
- (1)
- (60)
- (5)
- (5)
- (2)
- (4)
Related ORF Proteins:
Our products are intended for research use only, not for use in diagnostic, therapeutic or any other purposes.
Related Information and Products
SARS-CoV-2 Neutralizing Antibodies based on CR3022
SARS-CoV Protein Interactome / poster and products
SARS-CoV-2 RT-PCR Kits and qPCR Kits
SARS-CoV-2 Life Cycle: Stages and Inhibition Targets
Coronavirus Expression Vectors and Cloning Vectors at genomics-online.com.
References
: "SARS-CoV-2 Disrupts Splicing, Translation, and Protein Trafficking to Suppress Host Defenses." in: Cell, Vol. 183, Issue 5, pp. 1325-1339.e21, (2020) (PubMed).: "Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19." in: Cell, Vol. 181, Issue 5, pp. 1036-1045.e9, (2020) (PubMed).
: "The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade." in: Antiviral research, Vol. 176, pp. 104742, (2020) (PubMed).
: "Neuropilin-1 is a host factor for SARS-CoV-2 infection." in: Science (New York, N.Y.), Vol. 370, Issue 6518, pp. 861-865, (2020) (PubMed).
: "Host Factors in Coronavirus Replication." in: Current topics in microbiology and immunology, Vol. 419, pp. 1-42, (2019) (PubMed).
: "SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor." in: Cell, Vol. 181, Issue 2, pp. 271-280.e8, (2020) (PubMed).
: "Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant." in: Journal of translational medicine, Vol. 18, Issue 1, pp. 179, (2020) (PubMed).
: "COVID-19 infection: the perspectives on immune responses." in: Cell death and differentiation, Vol. 27, Issue 5, pp. 1451-1454, (2020) (PubMed).
: "C-reactive protein correlates with computed tomographic findings and predicts severe COVID-19 early." in: Journal of medical virology, Vol. 92, Issue 7, pp. 856-862, (2020) (PubMed).
: "The trinity of COVID-19: immunity, inflammation and intervention." in: Nature reviews. Immunology, Vol. 20, Issue 6, pp. 363-374, (2020) (PubMed).
: "Severe COVID-19: NLRP3 Inflammasome Dysregulated." in: Frontiers in immunology, Vol. 11, pp. 1580, (2020) (PubMed).
: "A single-cell atlas of the peripheral immune response to severe COVID-19." in: medRxiv : the preprint server for health sciences, (2020) (PubMed).
: "Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation." in: Science (New York, N.Y.), Vol. 367, Issue 6483, pp. 1260-1263, (2020) (PubMed).
: "Inflammasomes and Pyroptosis as Therapeutic Targets for COVID-19." in: Journal of immunology (Baltimore, Md. : 1950), Vol. 205, Issue 2, pp. 307-312, (2020) (PubMed).
: "Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors." in: Science (New York, N.Y.), Vol. 368, Issue 6489, pp. 409-412, (2020) (PubMed).
: "The CD14+ CD16+ blood monocytes: their role in infection and inflammation." in: Journal of leukocyte biology, Vol. 81, Issue 3, pp. 584-92, (2007) (PubMed).
- Diao, B. et al. Reduction and Functional Exhaustion of T Cells in Patients With Coronavirus Disease 2019 (COVID-19). Front. Immunol. 11, 827 (2020). Link to frontiersin.org
- Gao, T. et al. Highly pathogenic coronavirus N protein aggravates lung injury by MASP-2- mediated complement over-activation. medRxiv (2020). doi:10.1101/2020.03.29.20041962 Link to medRxiv
- Gao, Y. et al. Structure of RNA-dependent RNA polymerase from 2019-nCoV, a major antiviral drug target. bioRxiv 2020.03.16.993386 (2020). doi:10.1101/2020.03.16.993386 Link to bioRxiv
- Gordon, D. E. et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature (2020). doi:10.1038/s41586-020-2286-9 Link to nature.com
- Zhang, D. et al. COVID-19 infection induces readily detectable morphological and inflammation-related phenotypic changes in peripheral blood monocytes, the severity of which correlate with patient outcome. medRxiv (2020). doi:10.1101/2020.03.24.20042655 Link to medRxiv

Goal-oriented, time line driven scientist, proficiently trained in different academic institutions in Germany, France and the USA. Experienced in the life sciences e-commerce environment with a focus on product development and customer relation management.
Go to author page



