Epigenetics
Epigenetics is defined as heritable changes in gene expression that are, unlike mutations, not attributable to alterations in the sequence of DNA. The predominant epigenetic mechanisms are DNA methylation, modifications to chromatin, loss of imprinting and non-coding RNA. Epigenetic regulation of gene expression appears to have long-term effects and wide-ranging effects on health.1 There is a clear and prescient implication of benefits to both basic science and human health research that stems from a more complete understanding of the epigenome. These benefits have driven researchers to flock this field, and funding organizations have followed suit.
A large part of epigenetics is controlled by post-translational modifications on proteins. Methylations, acetylations, ubiquitination or phosphorylations lead to changes that for example influence transcription. Epigenetic modifications happen often during an organism’s lifetime; however, these changes can be transferred to the next generation if they occur in germ cells. Paramutation, bookmarking, imprinting, gene silencing, X chromosome inactivation, position effect, changeable disorder or phenotypic severity, reprogramming, maternal attributes, carcinogenic processes, teratogenic effects, regulation of histone modifications, heterochromatin states and cloning are known to involve epigenetic processes.2
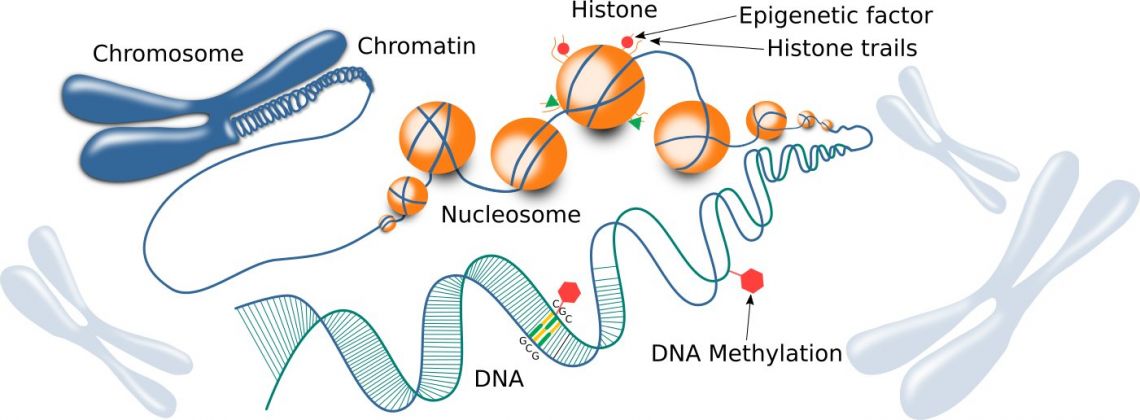
Fig. 1: Chromatin structure, including histones and DNA.
DNA Modifications
Covalent attachment of a methyl group to the C5 position of cytosine comprises the principal epigenetic modification of DNA. The presence of a methylated cytosine can repress transcription by inhibiting the binding of transcription factors or may promote the binding of other transcriptional repressors, including histone-modifying proteins, such as histone deacetylases.3
DNA demethylation plays an important role in development and tumorigenesis in mammals. DNA demethylation, occurring in primordial germ cells and in early embryos, is essential for cells to return to a pluripotent state.4
Histone Modifications
Similarly to DNA methylation, posttranslational histone modifications do not affect DNA nucleotide sequence but can modify its availability to the transcriptional machinery. Several types of histone modifications are known, amongst which acetylation, methylation, phosphorylation, and ubiquitination are the best studied and most important in terms of the regulation of chromatin structure and (transcriptional) activity. 5 While DNA methylation is considered to be a very stable epigenetic modification, histone modifications are comparably more labile with their levels being maintained by a balance between histone modifying enzymes, which add or remove specific modifications. Aberrant histone modification levels result from an imbalance in these modifying enzymes in diseased tissue, thus correcting the increased or decreased level of a particular enzyme will restore the natural balance in the affected cells.6
Chromatin Remodeling
The presence of histones acts as a barrier to protein access; thus chromatin remodeling must occur for essential processes such as transcription and replication. In conjunction with histone modifications, DNA methylation plays critical roles in gene silencing through chromatin remodeling. Chromatin remodeling is also interconnected with the DNA damage response, maintenance of stem cell properties, and cell differentiation programs. Chromatin modifications have increasingly been shown to produce long-lasting alterations in chromatin structure and transcription. Recent studies have shown environmental exposures in utero have the potential to alter normal developmental signaling networks, physiologic responses, and disease susceptibility later in life during a process known as developmental reprogramming.7
Transcription Factors
Chemical modifications of histones and DNA, such as histone methylation, histone acetylation, and DNA methylation, play critical roles in epigenetic gene regulation. Many of the enzymes that add or remove such chemical modifications are known, or might be suspected, to be sensitive to changes in intracellular metabolism. They (often) utilize cosubstrates generated by cellular metabolism, thereby providing a potential link between nutrition, metabolism, and gene regulation.8
Popular Epigenetics Antibodies
- (4)
- (21)
- (1)
- (8)
- (14)
- (1)
- (7)
- (20)
- (1)
- (1)
- (9)
- (1)
- (4)
- (1)
- (4)
- (5)
- (1)
- (3)
- (5)
- (1)
- (1)
- (1)
- (1)
- (3)
Need Help? Call our PhD Customer Support!
- We help you with finding the right product for your research.
- We offer reliable antibodies, kits, proteins, lysates for your research.
- Contact us via email or phone: (877) 302 8632 (US) or +49 241 95 163 153 (International)
Related Information and Products
Introduction to Chromatin immunoprecipitation (ChIP)
CUT&RUN - Improved Method to Study Protein-DNA Interactions
References
- : "Epigenetics: principles and practice." in: Digestive diseases (Basel, Switzerland), Vol. 29, Issue 2, pp. 130-5, (2011) (PubMed).
- : "Role of Epigenetics in Biology and Human Diseases." in: Iranian biomedical journal, Vol. 20, Issue 5, pp. 246-58, (2017) (PubMed).
- : "Epigenetic modifications: basic mechanisms and role in cardiovascular disease (2013 Grover Conference series)." in: Pulmonary circulation, Vol. 4, Issue 2, pp. 169-74, (2014) (PubMed).
- : "DNA methylation: superior or subordinate in the epigenetic hierarchy?" in: Genes & cancer, Vol. 2, Issue 6, pp. 607-17, (2011) (PubMed).
- : "Histone modifications and their role in epigenetics of atopy and allergic diseases." in: Allergy, asthma, and clinical immunology : official journal of the Canadian Society of Allergy and Clinical Immunology, Vol. 14, pp. 39, (2018) (PubMed).
- : "Epigenetic modifications as therapeutic targets." in: Nature biotechnology, Vol. 28, Issue 10, pp. 1069-78, (2015) (PubMed).
- : "Long-term effects of chromatin remodeling and DNA damage in stem cells induced by environmental and dietary agents." in: Journal of environmental pathology, toxicology and oncology : official organ of the International Society for Environmental Toxicology and Cancer, Vol. 32, Issue 4, pp. 307-27, (2014) (PubMed).
- : "Influence of metabolism on epigenetics and disease." in: Cell, Vol. 153, Issue 1, pp. 56-69, (2013) (PubMed).




